
Engineering the Neural Microenvironment

We use engineering approaches to develop 1) new therapies that promote regeneration of brain and spinal cord tissues and 2) improved preclinical models of brain tumors, and 3) ex vivo models of neuronal circuitry involved in pathologies.
A common theme across many pathologies of central nervous system, including cancer and spinal cord injury, is significant alterations to the extracellular matrix, such as changes in biochemical composition and structural mechanics of the tissue. A lack of understanding of how the extracellular matrix is fundamentally altered by injury or disease, and thus how these changes may be reversed to restore normal function, has been a major limitation to the development of new treatments. My lab develops tools for investigating the unique extracellular environment in the central nervous system and its relationship to cell function. We use biomimetic, hydrogel-based microenvironments with modular features that can be tuned to quantify independent effects of various extracellular parameters over cell phenotype and behavior and, in turn, provide a means for precise external control over cell and tissue function. Furthermore, we are developing methods for high-throughput assays of live cells cultured within these same biomaterial platforms. Acquisition of dynamic, high-throughput data from cells cultured in physiologically relevant conditions will provide new insights into pathologies of the central nervous system and, ultimately, more effective therapies for these conditions.
Research
Glioblastoma Tumor Microenvironment
Glioblastoma (GBM) is a highly aggressive type of cancer that originates in the brain and rarely metastasizes. The unique microenvironment of brain tumors is thought to be largely responsible for the characteristic aggressiveness and resistance to conventional treatments observed in GBM. However, the mechanisms underlying this relationship, are largely unknown. My lab works to develop physiologically translatable models that can be used to study how specific interactions between cells and the extracellular matrix in the microenvironment affect tumor physiology. We are engineering hydrogels as ex vivo culture systems based on hyaluronic acid (HA), a matrix polysaccharide abundant in the brain, to mimic the various aspects of the tumor microenvironment. HA and its cell surface receptors are dramatically upregulated near GBM tumors and this shift likely has a substantial contributes to the increased the migratory capacity and resistance to chemotherapeutic drugs. GBM cells cultured within these 3D environments more closely mirror their in vivo counterparts than those cultured using standard methodologies. Precise and independent control of the biochemical and biophysical landscape in these culture platforms enables investigation of the quantitative effects of these features on cell physiology so that clinically relevant data can be acquired in an ex vivo setting. Our research aims to characterize the microenvironment of clinical tumors and engineer culture planforms for patient-derived GBM cells that recapitulate these features. We then apply these cultures to better understand tumor physiology and develop new, patient-tailored treatment strategies.
Brain and Spinal Cord Regeneration

Biomaterial Scaffolds
A major focus of our lab is developing strategies to regenerate brain and spinal cord tissues; for example, after injury or tumor resection. Clinically effective strategies to restore nerve tissues will require consideration multiple inherent barriers, such as formation of a chronic inflammatory microenvironment and a limited activity of endogenous stem cells. Biomaterials can be used to deliver coordinated, combinatorial therapies that include an extracellular matrix-like substrate, pro-regenerative biomolecules, electrical stimulation and cell transplants. Hydrogels are ideal biomaterials for use in the central nervous system because they can be injected and formed directly in living tissue, approximate mechanical and biochemical properties of healthy tissue and deliver regenerative therapies including cells. We are developing hydrogels based on hyaluronic acid (HA), a high molecular weight, matrix polysaccharide, reported to significantly augment wound healing, nerve regeneration and cell migration. We are working to maximize tissue regeneration by optimizing architecture, mechanical properties and biological signals of these biomaterial scaffolds.
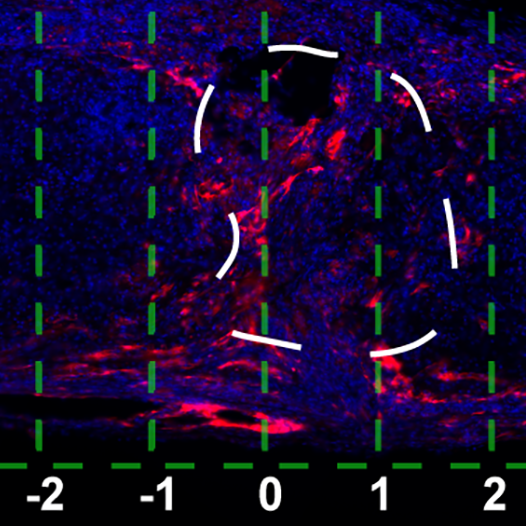
Biomaterial-Mediated Gene Delivery
Delivery of genetic vectors from biomaterials is a powerful tool to influence the in vivo microenvironment. Lentiviral vectors enable sustained, localized protein expression and facile delivery of virtually any factor and combinations of factors simply by encapsulation of viral particles within hydrogels. We aim to engineer hydrogel scaffolds that support delivery of lentiviral vectors encoding for regenerative factors and to evaluate their effects on regeneration of the brain and spinal cord.
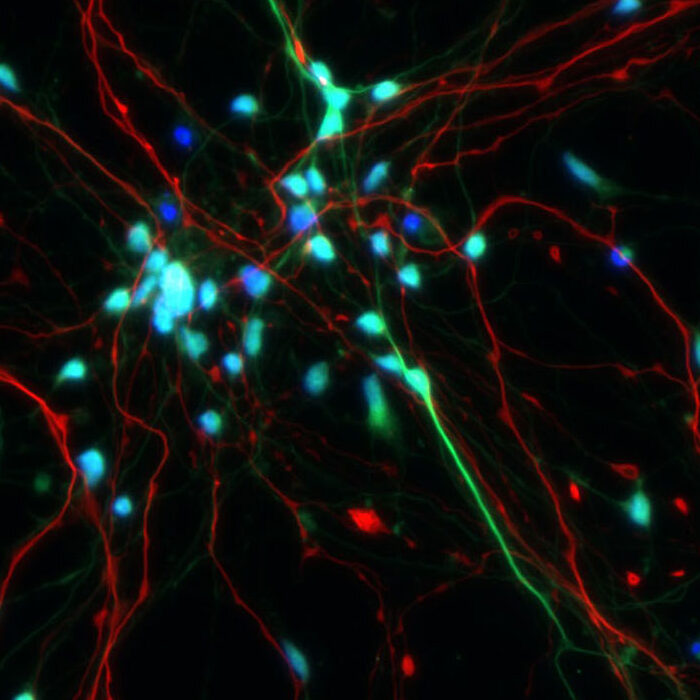
Stem Cell Transplants
Therapeutic delivery of neural stem and progenitor cells (NSPCs) has shown promise to treat restore neurological functions lost to injury and disease. However, low rates of survival, appropriate differentiation and functional engraftment after transplantation have been major limitations. To address these issues, we are working to develop hydrogel scaffolds that can provide NSPCs with 1) localized delivery, 2) improved survival during injection, 3) a supportive scaffolding after transplantation, 4) shielding from an inflammatory microenvironment, 5) factors that actively promote differentiation and 6) an architecture promoting integration with host tissue.
Microphysiological Models of the Central Nervous System
Because of our limited understanding of the highly complex cell-cell and cell-matrix interactions in the central nervous system and how these interactions coordinate tissue function, in vitro experimental models have not adequately reflected in vivo physiology necessitating intensive use of animal models. Biomaterials can be engineered to present defined, extracellular matrices that can greatly simplify the experimental variability in biological systems so that effects of a single variable can be experimentally isolated in vitro. We are culturing human neural stem cells within such 3D, biomaterial-based platforms to identify key physiological interactions between cells and their environment during central nervous system tissue development, repair, and disease progression. In addition, we have developed methods for high-throughput screening within these biomimetic cultures that can be used for development of new clinical therapies.
People

Principal Investigator
Stephanie Seidlits, Ph.D.
Postdoctoral Research, Northwestern University
(Chemical & Biological Engineering)
Ph.D., M.S. University of Texas, Austin
(Biomedical Engineering)
B.S., Rice University (Bioengineering)
Research Areas: cell and tissue engineering, neural tissue engineering, spinal cord injury, glioblastoma multiforme, extracellular matrix, hyaluronic acid, biomaterials, hydrogels, signal transduction, lentiviral vectors, gene therapy

Lab Manager
Zheng Ma, Ph.D.
Postdoctoral Research, University of Texas Southwestern Medical Center
(mechanisms of transcriptional elongation and co-transcriptional splicing)
Ph.D., University of Toledo (Biochemistry)
B.S., Shenyang Agricultural University
(Bioengineering)

Senior Research Fellow
Manish K. Jaiswal, Ph.D.
manish.jaiswal@austin.utexas.edu
Research Scientist, Postdoctoral Research, Texas A&M University
(biomaterials, nanomaterials, tissue engineering)
Ph.D., Indian Institute of Technology, Bombay (Biomaterials)
M.S., Banaras Hindu University, Banaras
Research areas: biomaterials, tissue engineering

Post-Doctoral Researcher
Niraj Babu, Ph.D.
Ph.D., Institute of Bioinformatics, Bangalore (Oncology and Cancer Biology)
M.S., National Institute of Technology Rourkela (Biotechnology)
B.Tech., Sir M Visvesvaraya Institute of Technology (Biotechnology)
Research areas: glioblastoma, cancer metabolism, therapyresistance, signal transduction

Post-Doctoral Researcher
Hyuna Kim, Ph.D.
Ph.D., University of Massachusetts, Amherst
B.S., Korea University
(Biotechnology, Medical Science, dual B.S.)
Research Areas: biomaterials, glioblastoma, neural interactions

Post-Doctoral Researcher
Alireza Sohrabi, Ph.D.
alireza.sohrabi@austin.utexas.edu
Ph.D., University of California, Los Angeles (Bioengineering)
M.S., University of Twente (Chemical Engineering)
B.S., Amirkabir University of Technology, Tehran Polytechnic (Polymer Chemistry)
Research Areas: glioblastoma, biomaterials, cell migration, cancer metabolism, mechanotransduction

Post-Doctoral Researcher
Ze Zhong (Bill) Wang, Ph.D.
zezhong.wang@austin.utexas.edu
M.S., Ph.D., Washington University, St. Louis (Biomedical Engineering)
B.S., University of California, San Diego (Bioengineering)
Research areas: brain-on-a-chip models, neural stem cells, biomaterials, nigrostriatal pathway

Ph.D. Student Researcher
Rebecca Bierman Duquette, M.S.
M.S., University of California, Los Angeles (Bioengineering)
B.S., Georgia State University (Neuroscience)
Research areas: spinal cord injury, conductive hydrogels, neural stem cell therapies

Ph.D. Student Researcher
Mollie Harrison
molliejharrison@utmail.utexas.edu
B.S., University of Missouri-Columbia
(Chemical Engineering)
Research Areas: glioblastoma, therapy resistance, signal transduction, biomaterials

Ph.D. Student Researcher
Chris Houk
B.S., Texas A&M University
(Biomedical Engineering)
Research Areas: biomaterials, 3D printing

Ph.D. Student Researcher
Shabnam Nejat
shabnam.nejat@utmail.utexas.edu
B.S., M.S., Saint Louis University
(Biomedical Engineering)
Research Areas: biomaterials, glioblastoma

Ph.D. Student Researcher
Sabrina Pietrosemoli Salazar
B.S., North Carolina State University - University of North Carolina at Chapel Hill
(Biomedical and Health Science Engineering)
Research areas: neural stem cells, CNS injury, 3D culture

Ph.D. Student Researcher
Talia Sanazzaro
talia.sanazzaro@utmail.utexas.edu
B.S., Washington State University (Bioengineering)
Research areas: glioblastoma, biomaterials, co-cultures, cancer vasculature

Ph.D. Student Researcher
Breahna Singer
B.S., University of California at Los Angeles (Physiological Sciences)
Research areas: glioblastoma, biomaterials, co-cultures, cancer vasculature
Undergraduate Student Researchers
-
Ademola Adetosoye
-
Neha Arvinth
-
Ananya Bharadwaj
-
Reyan Ghanim
-
Adhishree Luintel
-
Sahana Murthy
-
Arushi Nath
-
KhanhVy Nguyen
-
Morgan Owens
-
Amy Poe
-
Rohan Srinivas
-
Nadia Toh
Past Lab Members
Postdoctoral Researchers
Soniya Bastola, Ph.D. (UCLA, 2022)Jessica Chen, Ph.D. (UCLA, 2022)
Christopher Walthers, Ph.D. (UCLA, 2017)
Ph.D. Student Researchers
Arshia Ehsanipour, Ph.D. (UCLA, 2019)Jesse Liang, Ph.D. (UCLA, 2022)
Joshua Karam, Ph.D. (UCLA, 2022)
Alireza Sohrabi, Ph.D. (UCLA, 2021)
Weikun Xiao, Ph.D. (UCLA, 2019)
M.S. Student Researchers
Rebecca Bierman, M.S. (UCLA, 2018)Bernardo Maximiliano Garduño, M.S. (UCLA, 2019)
Alexis Morrison Litke, M.S. (UCLA, 2018)
Magdalena Morysewicz, M.S. (UCLA, 2015)
Bushra Rajput, M.S. (UCLA, 2022)
Gevick Safarians, M.S. (UCLA, 2022)
Undergraduate Student Researchers
Valeria Virrueta (2022, UT Austin, UT BUILDING Scholar)Tasha Aboufadel (2016, UCLA)
Oyindamola Akinnusi (2019, UCLA)
Eunice Alvarado (2020, UCLA)
Aishani Ataliwala (2016, UCLA)
Christopher Anisi (2022, UCLA)
Mabel Chen (2020, UCLA)
Peter Cho (2017, UCLA, Amgen Scholars Summer Research Program)
Phillip Cox (2017, UCLA)
Robel Dagnew (2016, UCLA)
Mary Epperson (2022, UCLA)
Sohib Ezubeik (2021, UCLA)
Sarahi Garza (2020, UCLA)
Elnaz Guivatchian (2020, UCLA)
Michael Hashimoto (2018, UCLA)
Jeremy Huang (2021, UCLA)
Carolyn Kim (2018, UCLA)
Tiffany Kyu (2017, UCLA)
Amelia Lao (2018, UCLA)
Qingyang Li (2017, UCLA)
Sihan (Venus) Liu (2018, UCLA)
Peipei Lyu (2017, UCLA)
Kajal Maran (2020, UCLA)
Jaimie Mayner (2016, UCLA)
Nadia Mortezagholi (2017, UCLA)
Tommy Nguyen (2018, UCLA)
Bianca Onyekwere (2019, UCLA)
Nico Pedroncelli (2021, UCLA)
Laila Rad (2020, UCLA)
Sara Rashidi (2017, UCLA)
Isabella Rosenzweig (2022, UCLA)
Maylione Sathialingam (2021, UCLA)
Savannah Shumock (2020, UCLA)
Itay Solomon (2020, UCLA)
Ryan Stoutamore (2017, UCLA)
Catherina (Songping) Sun (2017, UCLA)
Kelly Tamura (2022, UCLA)
Zachary Tom (2018, UCLA)
Ryan Tsang (2019, UCLA)
Joseph Tsung (2017, UCLA)
Shanshan Wang (2019, UCLA)
Alexander Wu (2016, UCLA)
Xiancheng (Lewis) Wu (2015, UCLA)
Julius Yee (2017, UCLA)
Wutong Yu (2019, UCLA)
Meagan Yuen (2016, UCLA)
Rongyu Zhang (2018, UCLA)
Contact

Department of Biomedical Engineering
University of Texas at Austin
Engineering Education and Research Center
Austin, TX 78712
seidlits@utexas.edu
zheng.ma@austin.utexas.edu
Publications
- 1. S.R. Peyton, L.W. Chow, S.D. Finley, A.N. Ford Versypt, R. Hill, M.L. Kemp, E.M. Langer, A.P. McGuigan, A.S. Meyer, S.K. Seidlits, K. Roy, and S.M. Mumenthaler. (2023) Synthetic living materials in cancer biology. Nature Reviews Bioengineering. 1, 972-988. doi: 10.1038/s44222-023-00105-w.
- 2. S. Bastola, M.S. Pavlyukov, Y. Ghochani, N. Sharma, M.A. Nakano, S.D. Muthukrishnan, S.Y. Yu, M.S. Kim, A. Sohrabi, N.P. Biscola, D. Yamashita, K.S. Anufrieva, R. Kawaguchi, Y. Qin, S.K. Seidlits, A.L. Burlingame, J.A. Oses-Prieto, L.A. Havton, S.A. Goldman, A.B. Hjelmeland, I. Nakano, H.I. Kornblum. (2023) Endothelial-secreted Endocan protein acts as a PDGFR alpha ligand and regulates vascularity, radioresistance, and regional phenotype in glioblastoma. Biorxiv, 2023 Sept. 8. doi: 10.1101/2020.10.12.335091. Preprint.
- 3. A.J. Anderson, U. Nekanti, P. Sakthivel, A. Zahedi, D.A, Creasman, R. Nishi, C.M. Dumont, K. Piltti, G.L. Guardamondo, N. Hernandez, X. Chen, H. Song, X. Lin, J. Martinez, L. On, A. Lakatos, K. Pawar, B. David, Z. Guo, S.K. Seidlits, X. Xu, L.D. Shea, B.J. Cummings. (2023) Multichannel bridges and NSC synergize to enhance axon regeneration, myelination, synaptic reconnection, and recovery after SCI. Res Sq. 2023 Jul 19;rs.3.rs-3044426. doi: 10.21203/rs.3.rs-3044426/v1. Preprint. NPJ Regenerative Medicine. Accepted and in press, 2024.
- 4. A. Sohrabi, A.E.Y.T. Lefebre, M.J. Harrison, M.C. Condro. T.M. Sanazzaro, G. Safarians, I. Solomon, S. Bastola, S. Kordbacheh, N. Toh, H.I. Kornblum, M.A. Digman, S.K. Seidlits. (2023) Microenvironmental stiffness induces metabolic reprogramming in glioblastoma. Cell Reports Oct 31;42(10):113175. Doi: 10.1016/j.celrep.2023.113175.
- 5. J. Karam, B.J. Singer, H. Miwa, L.H. Chen, K. Maran, M. Hasani, S. Garza, B. Onyekwere, H.C. Yeh, S. Li, D. Di Carlo, S.K. Seidlits. (2023) Molecular weight of hyaluronic acid crosslinked into biomaterial scaffolds affects angiogenic potential. Acta Biomaterialia. 169:228-242. doi: 10.1016/j.actbio.2023.08.001.
-
G. Safarians*, A. Sohrabi*, I. Solomon*, W. Xiao, S. Bastola, B.W. Rajput, M. Epperson, I. Rosenzweig, K. Tamura, B. Singer, J. Huang, M.J. Harrison, T. Sanazzaro, M.C. Condro, H.I. Kornblum, S.K. Seidlits. (2023) Glioblastoma cell invasion through soft, brain-like matrices depends on tethering of extracellular hyaluronic acid to the actin cortex through CD44 and ezrin. Advanced Healthcare Materials. 12(14): 2203143. doi: 10.1002/adhm.202203143.
-
Z.Z. Khaing*, J.Y. Chen, G. Safarians, S. Ezubeik, N. Pedroncelli, R.D. Duquette, S.K. Seidlits*. *co-corresponding authors (2023) Clinical trials targeting secondary damage after traumatic spinal cord injury. Int. J. Mol. Sci. 24(4):3824. doi: 10.3390/ijms24043824.
-
W. Xiao, M. Pahlavan, C.Y. Eun, X. Zhang, C. DeKalb, B. Mahgoub, H. Knaneh, S. Shah, A. Sohrabi, S.K. Seidlits, R. Hill. (2022) Matrix stiffness mediates pancreatic cancer chemoresistance through induction of exosome hypersecretion in cancer associated fibroblasts. Matrix Biol. Plus. 14:100111, doi: 10.1016/j.mbplus.2022.100111.
-
L. Amirifar, A. Shamlo, R. Nasiri, N.R. de Barros, Z.Z. Wang, B.D. Unluturk, A. libanori, O. Ievglevskyi, S.E. Diltemiz, S. Sances, I. Balasingham, S.K. Seidlits*, N. Ashammakhi*. *co-corresponding authors (2022) Brain-on-a-chip: Recent advances in design, biomaterials, and cell culture techniques for microfluidic models of the brain in health and disease. Biomaterials. 285:121531.
-
J. Liang, A. Sohrabi, M. Epperson, L. Rad, M. Sathialingam, P. Lue, J. Huang, J. Popoli, A. Yackley, M. Bick, T. Skandakumar, Z.Z. Wang, C.C. Chen, G. Varuzhanyan, R. Damoiseaux, S.K. Seidlits. (2022) Hydrogel arrays enable increased throughput for screening effects of matrix components and therapeutics in 3D tumor models. J. Vis. Exp. (JoVE). Published online June 2022. doi: 10.3791/63791.
-
Y. Ghochani, A. Sohrabi, S.D. Muthukrishnan, R. Kawaguchi, M.C. Condro, S. Bastola, F. Gao, Y. Qin, J. Motttahedeh, M.L. Iruela-Arispe, N. Rao, D.R. Laks, L.M. Liau, G.W. Mathern, S.A. Goldman, S.T. Carmichael, I. Nakano, G. Coppola, S.K. Seidlits*, H.I. Kornblum* (co-corresponding authors*). (2022) Generation of a molecular interactome of the glioblastoma perivascular niche reveals Integrin Binding Sialoprotein as a key mediator of tumor cell migration. Cell Reports, published online Nov. 2022; Biorxiv doi: 10.1101/2021.10.01.462643.
-
R.D. Bierman-Duquette, G. Safarians, J. Huang, B. Rajput, J.Y. Chen, Z.Z. Wang, S.K. Seidlits (2021) Engineering tissues of the central nervous system: Interfacing conductive biomaterials with neural stem/progenitor cells. Advanced Healthcare Materials 11(7):e2101577. doi: 10.1002/adhm.202101577.
-
S.K. Seidlits, K.A. Killian. (2021) Biomaterials for personalized disease models. Acta Biomater., 132:1-3.
-
S.E. Diltemiz, M. Tavafoghi, N.R. de Barros, M. Kanada, J. Heinämäki, C. Contag, S.K. Seidlits, N. Ashammakhi. (2021) Use of artificial cells as drug carriers. Mater. Chem. Front. 5:6672-6692.
-
Q. Yu, W. Xiao, A. Sohrabi, S. Sun, A. Sohrabi, J. Liang, S.K. Seidlits. (2021) Extracellular matrix proteins confer cell adhesion-mediated drug resistance through integrin v in glioblastoma cells. Front. Cell Develop. Biol., 9:240. doi: 10.3389/fcell.2021.616580.
-
A. Ehsanipour, M. Sathialingam, L.M. Rad, J. de Rutte, R.D. Bierman, J. Liang, W. Xiao, D. Di Carlo, S.K. Seidlits. (2021) Injectable, macroporous scaffolds for delivery of therapeutic genes to the injured spinal cord. APL Bioengineering, 5(1):016104. doi: 10.1063/5.0035291. Featured article.
-
Bastola S., Pavlyukov M.S., Ghochani Y., Muthukrishnan S.D., Cho H.J., Lee Y., Sohrabi A., Seidlits, S.K., Yamashita D., Kim M.S., Anufrieva K.S., Kawaguchi R., Qin Y., Burlingame A.L., Oses-Prieto J.A., Goldman S.A., Hjelmeland A.B., Nam D., Kornblum H.I. & Nakano I. (2020) Tumor edge architecture in glioblastoma is constructed by inter-cellular signals from vascular endothelial cells. Biorxiv doi.org/10.1101/2020.10.12.335091.
-
A. Kita, J. Saldate, C. Chang, N. Chellappa, J. Jong, R. Matsuda, A. Schmidt, B. Shih, Ir. Shafqat, K. Schoettler, S. Acharaya, S. Seidlits, L. Hoffman. (2020) Implantable drug-eluting scaffolds as programmable reservoirs for inner ear delivery of pharmacotherapeutics. Otolaryngology-Head and Neck Surgery, 163(4):791-798.
-
A. Ehsanipour, T. Nguyen, T. Aboufadel, M. Sathialingam, P. Cox, W. Xiao, C.M. Walthers, S.K. Seidlits. (2019) Injectable, hyaluronic acid-based scaffolds with macroporous architecture for gene delivery. Cell. Mol. Bioeng., 12(5):399-413. *Special Issue: 2019 Young Innovators
-
W. Xiao, S. Wang, R. Zhang, A. Sohrabi, Q. Yu, S. Liu, A. Ehsanipour. J. Liang, R.D. Bierman, D.A. Nathanson, S.K. Seidlits. (2019) Bioengineered scaffolds for 3D culture demonstrate extracellular matrix-mediated mechanisms of chemotherapy resistance in glioblastoma. Matrix Biol., pii: 0945-053X(19):30022-30028. *Special Issue: Matrix Biomechanics
-
S.K. Seidlits (corresponding author), J. Liang, R.D. Bierman, A. Sohrabi, J. Karam, S. Holley, C. Cepeda, C.M. Walthers. (2019) Peptide-modified, hyaluronic acid-based hydrogels as a 3D culture platform for neural stem/progenitor cell engineering. J. Biomed. Mater. Res. Part A, 107(4):704-718. *selected for 2019 Society for Biomaterials Young Investigator Award
-
N. Ashammakhi*, H.J. Kim, A. Ehsanipour, R.D. Bierman, O. Kaarela, C. Xue, A. Khademhoesseni*, S.K. Seidlits*. *co-corresponding authors (2019) Regenerative therapy for spinal cord injury. Tissue Eng: Part B, 25(6):471-491.
-
W. Xiao, R. Zhang, S. Sun, A. Sohrabi, A. Ehsanipour, C. Walthers, J. Liang, R. Xu, L. Ta, D.A. Nathanson, S.K. Seidlits. (2018) Brain-mimetic, 3D culture platforms for investigating cooperative effects of extracellular matrix features on treatment resistance in glioblastoma. Cancer Res. 17(5):1758-70.
-
W. Xiao, A. Sohrabi, S.K. Seidlits. (2017) Integrating the glioblastoma microenvironment into engineered experimental models. Future Sci OA, 3(3), doi: 10.4155/fsoa-2016-0094.
-
Z.Z. Khaing, A.A. Ehsanipour, C.P. Hofstetter, S.K. Seidlits. (2016) Injectable hydrogels for spinal cord repair: A focus on swelling and intraspinal pressure. Cells Tissues Organs 202(1-2):67-84.
-
D.J. Margul, J. Park, R.M. Boehler, D.R. Smith, M.A. Johnson, D.A. McCreedy, T. He, A. Ataliwala, T.V. Kukushliev, Jesse Liang, Alireza Sohrabi, A.G. Goodman, C.M. Walthers, L.D. Shea*, S.K. Seidlits* (co-corresponding authors). (2016) Reducing neuroinflammation by delivery of IL-10 encoding lentivirus from multiple-channel bridges. Bioeng. & Transl. Med. 1(2):136-148.
-
B.P. Bernabé, S. Shin, P.D. Rios, L.J. Broadbelt*, L.D. Shea*, S.K. Seidlits* (co-corresponding authors). (2016) Dynamic transcription factor activity networks in response to independently altered mechanical and adhesive microenvironmental cues. Integrat. Biol. 8(8):844-60.
-
J. Lim, A. Ehsanipour, J.J. Hsu, J. Lu, T. Pedego, A. Wu, C.M. Walthers, L.L. Demer, S.K. Seidlits, Y. Tintut. (2016) Inflammation drives retraction, stiffening and nodule formation via cytoskeletal machinery in a 3-dimensional culture model of aortic stenosis. Am. J. Pathol., 186(9):2378-89.
-
M. Skoumal, S.K. Seidlits, S. Shin, L.D. Shea. Localized lentivirus delivery via peptide interactions. (2016) Biotech. Bioeng., 113(9):2033-40.
-
D.S. Hernandez, E.T. Ritschdorff, S.K. Seidlits, C.E. Schmidt, J.B. Shear. (2016) Functionalizing micro-3D-printed protein hydrogels for cell adhesion and patterning. J. Mater. Chem. B, 4:1818-1826.
-
D.A. McCreedy, D.J. Margul, S.K. Seidlits, R.M. Boehler, J. Antane, R. Thomas, D.R. Smith, T. He, T.V. Kukushliev, B. Vedia, J. Lamano, S.W. Goldsmith, G. Sissman, L.D. Shea. (2016) Semi-automated counting of axon regeneration in PLG spinal cord bridges. J. Neurosci. Methods, 263:15-22.
-
Z.Z. Khaing, S.K. Seidlits. (2015) Hyaluronic acid and neural stem cells: Implications for biomaterial design. J Mater Chem B, 3:7850-7866.
-
C.M. Walthers, S.K. Seidlits. (2015) Gene delivery strategies for spinal cord repair. Biomarkers Insights, Suppl 1: 11-29.
-
A.M. Thomas*, S.K. Seidlits*, (co-first authors*), A.G. Goodman, T.V. Kukushliev, D.M. Hassani, B.J. Cummings, A.J. Anderson, L.D. Shea. (2014) Sonic hedgehog and neurotrophin-3 increase oligodendrocyte numbers and myelination after spinal cord injury. Integrative Biology 6(7):694-705.
-
S.K. Seidlits, K.A. Hlavaty, L.D. Shea. (2014) “DNA delivery for regeneration” in Biomaterials and Regenerative Medicine, ed. P.X. Ma. Cambridge University Press, Cambridge, MA.
-
A.M. Thomas, M.B. Kubilius, S.J. Holland, S.K. Seidlits, R.M. Boehler, A. J. Anderson, B.J. Cummings, L.D. Shea. (2013) Channel density and porosity of degradable bridging scaffolds on axon growth after spinal injury. Biomaterials 34(9):2213-2220.
-
S.K. Seidlits, R.M. Gower, J.A. Shepard, L.D. Shea. (2013) Hydrogels for lentiviral gene delivery. Expert Opinion on Drug Delivery, 10(4):499-509.
-
Z.Z. Khaing, B.D. Milman, J.E. Vanscoy, S.K. Seidlits, R.J. Grill, C.E. Schmidt. (2011) High MW hyaluronic acid limits astrocyte proliferation and scar formation after SCI. J Neural Eng. 8(4):046033.
-
S.K. Seidlits, C.T. Drinnan, R.R. Petersen, J.B. Shear, L.J. Suggs, C.E. Schmidt. (2011) Fibronectin-hyaluronic acid composites for three-dimensional endothelial cell culture. Acta Biomaterialia 7(6):2401-2409.
-
Y. Yang, S.K. Seidlits, M.M. Adams, Lynch VM, C.E. Schmidt, E.V. Ansyln, J.B. Shear. (2010) A highly selective low-background fluorescent imaging agent for nitric oxide. J American Chemical Society 132(38):13114-13116.
-
S.K. Seidlits*, Z.Z. Khaing*, (co-first authors*) R.R. Petersen, J.D. Nickels, J.E. Vanscoy, J.B. Shear†, C.E. Schmidt† (co-corresponding authors). (2010) The effects of hyaluronic acid hydrogels with tunable mechanical properties on neural progenitor cell differentiation. Biomaterials 31:3930-3940.
-
S.K. Seidlits, C.E. Schmidt*, J.B. Shear* (co-corresponding authors*). (2009) High-resolution patterning of hydrogels in three dimensions using direct-write photofabrication for cell guidance. Advanced Functional Materials 19:3543-3551.
-
G. Kijanka, R. Barry, H. Chen, E. Gould, S.K. Seidlits, J. Schmid, M. Morgan, D.Y. Mason, J. Cordell, D. Murphy. (2009) Defining the molecular target of an antibody derived from nuclear extract of Jurkat cells using protein arrays. Analytical Biochemistry 395(2):119-124.
-
S.K. Seidlits, J.Y. Lee, C.E. Schmidt. (2008) Nanostructured scaffolds for neural applications. Nanomedicine, 3(2):183-99.
-
S.K. Seidlits, N.A. Peppas. (2007) “Star polymers and dendrimers in nanotechnology and drug delivery” in Nanotechnology in Therapeutics: Current Technology and Applications, ed. Peppas, N.A., Hilt, J.Z., Thomas, J.B. Horizon Press, pp. 317-348.
-
F.K. Kasper, S.K. Seidlits, A. Tang, R.S. Crowther, D.H. Carney, M.A. Barry, A.G. Mikos. (2005) In vitro release of plasmid DNA from oligo(poly(ethylene glycol) fumarate) hydrogels. J Controlled Release. 104(3):521-539.
-
S. Seidlits, T. Reza, K.A. Briand, A.B. Sereno. (2003) Voluntary spatial attention benefits voluntary not reflexive saccadic eye movements. TheScientificWorld Journal; 3:881-902.
Copyright 2022 Seidlits Lab